Biotechnology: Principles and Processes
Definition of biotechnology which covers both traditional views and modern molecular biotechnology has been given by European Federation of Biotechnology (EFB). According to EFB , “Biotechnology is the integrated use of biochemistry, microbiology and engineering sciences in order to achieve technological application of the capabilities of microorganisms, cultured tissues/cells and parts there of” . Let us have an overview of the principles and processes of Biotechnology.
Principles of Biotechnology
According to modern Biotechnology, the main principles of Biotechnology are:
- Genetic engineering, which is used to modify the DNA of the target organism, thereby changing the phenotype of the organism.
- Bioprocess engineering, which is the maintenance of sterile conditions to support the growth of large quantities of desired microbes and other eukaryotic cells which are used for the production of new or modified biotechnological products such as antibiotics, enzymes, vaccines, etc.
The techniques of genetic engineering mainly include:
- DNA fragment is isolated from the donor organism.
- It is inserted into the vector DNA.
- It is transferred into an appropriate host.
- Cloning of the recombinant DNA in the host organism.
What is Recombinant DNA technology?
Recombinant DNA technology is also known as Genetic Engineering. It is the process of joining together two DNA molecules from two different organisms. This is known as the recombinant DNA.
The steps involved in the processes of Recombinant DNA technology are:
- Isolation of DNA
- DNA fragmentation using restriction endonuclease.
- Ligation of the desired DNA fragment into the vector.
- Transfer of the recombinant DNA into the host.
- Culture of the transformed cells in a nutrient medium.
- Extraction of the desired product.
DNA Cloning
DNA cloning is the process of making multiple, identical copies of a piece of DNA. This process requires cloning vectors which possess the following properties:
It should be smaller in size but should be able to carry a large DNA insert.
The cloning vector should have the origin of replication so that it can autonomously replicate in the host organism.
It should have a restriction site.
It should have a selectable marker to screen recombinant organism.
It should possess multiple cloning sites.
Basic Principles of Biotechnology:
Genetic engineering allows the isolation and introduction of only the desired genes into the organism without introducing the undesirable genes. The steps involved in genetic engineering are:
1. Development of recombinant DNA (rDNA).
2. Cloning of desired gene
3. Transfer of the cloned gene into suitable host organism.
➢ Origin of replication (ori):
A specific DNA sequence in the chromosome that can initiate DNA replication. The foreign DNA introduced into the host genome has to be linked to the origin of replication in the host chromosome for the gene to be able to multiply. If the foreign gene is not linked to the ori sequence it may not be able to multiply.
➢ Cloning:
The process of making multiple identical copies of a template DNA
➢ Plasmid:
A circular extra-chromosomal material that is capable of autonomous replication. Plasmids are used as vectors for cloning and expression. Foreign gene is introduced into a plasmid and the plasmid is allowed to multiply. This causes the multiplication of the desired gene.
➢ Antibiotic resistance gene:
The gene in certain microorganisms that bestows on them the ability to grow in the presence of the specific antibiotic as the gene gives them resistance. These genes are present on plasmids. These are used as indicators of cloning and transformation.
➢ Restriction Enzymes:
They are enzymes that can cut DNA at specific fragments. They are also called as “molecular scissors”. The sequences at which they cut the DNA are specific for the restriction enzyme. They allow the desired gene to be cut and be introduced in specific locations in the vector or host DNA.
➢ Vectors:
These are plasmids that are used to multiply and transfer the desired gene from one organism to the next.
➢ Ligase:
Enzymes that are responsible for the joining of the desired gene fragment with the host DNA. Ligases function by getting DNA fragments to stick together.
➢ The basic steps in genetic modification of an organism:
Identification of desired DNA fragment.
Introduction of desired DNA fragment into suitable host.
Maintaining foreign DNA in the host and its transfer to the progeny.
Tools for genetic engineering (Recombinant DNA Technology):
Restriction enzymes or molecular scissors are used to cut DNA to be inserted into the vector. These enzymes add methyl group to the DNA, which help in restricting the digestion of their own DNA. They are used to cut DNA fragments with specific recognition sequences.
Recognition sequences:
The sequence of DNA bases that can be recognized by the restriction enzyme as the site for restriction or cutting. They exist as palindromic sequences. Recognition site for EcoRI below—
 |
| EcoRI - Restriction Endonuclease |
There are two types of restriction enzymes- endonuclease and exonuclease.
Endonuclease cut the DNA in the middle whereas exonuclease cut at the ends. For example, ECoR1, Hind III, etc. are examples of restriction endonuclease. Restriction enzymes cut at a specific site on DNA known as restriction site. The restriction site is characterised by a specific recognition sequence for the endonuclease. Each restriction endonuclease identifies a specific palindromic nucleotide sequences in DNA. Palindrome in DNA is a sequence of base pairs that are present in the same order on the two strands when orientation of reading is kept same.
 |
| Action of Restriction endonuclease |
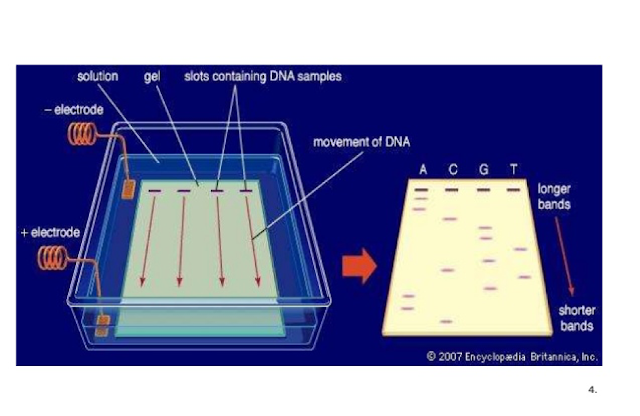
Ligases are the enzyme that joins the two DNA fragments. Presence of sticky ends (similar overhanging sequences due to the action of the same restriction enzyme) helps in ligation. Separation and Isolation of DNA Fragments: The DNA fragments obtained through restriction are separated by a technique called as gel electrophoresis.
 |
| DNA ligase |
Gel Electrophoresis: It involves the migration of negatively charged DNA to the positive electrode through a porous polymer gel matrix under the influence of an electric field. The DNA fragments separate or resolve depending on their size as well as the pore size of the gel. The smaller DNA fragments are able to migrate farther than the larger DNA fragments.
The most common matrix used for DNA electrophoresis is agarose. Agarose is obtained from seaweeds.
Visualisation: DNA fragments cannot be directly observed. To observe the DNA fragments they need to be first stained with a compound called as ethidium bromide (EtBr) and then placed in UV light. DNA stained with EtBr fluoresces under UV.
Elution: Purification of desired DNA fragments from the gel using various methods is called as elution.
Agarose Gel Electrophoresis of DNA :
 |
| Agarose Gel Electrophoresis |
Cloning vectors(Vehicle DNA or Carrier DNA)
The vectors are DNA molecules that can carry a foreign DNA segment and replicate inside the host cell. Vectors may be plasmids, bacteriophages, cosmids, yeast artificial chromosomes(YACs), bacterial artificial chromosomes (BACs) and viruses.
Following features are required for a cloning vector :
• Origin of replication, this is known as ori. This is important for replication within the host cell as well as to maintain the copy number.
• Selectable marker to identify transformed cells. Transformation is the process used to introduce piece of DNA into the host cells. The genes encoding resistance to antibiotics such as ampicillin, chloramphenicol, tetracycline, or kanamycin, etc. are some of the useful selectable markers for E. coli. The normal E. coli cells do not show any resistance against any of these antibiotics.
• There should be cloning site in the cloning vector. Presence of more than one recognition site can complicate the cloning, so single cloning site is preferable. The ligation of the foreign DNA usually occurs at the site of antibiotic resistance gene. Once of the gene of interest gets inserted at the site of antibiotic resistance gene, antibiotic resistance will be lost. So, a recombinant plasmid will lose antibiotic resistance. So, recombinants can be selected from the non-recombinants. Another method to find out the transformed cells is insertional inactivation. This is based on the ability to produce color in the presence of a chromogenic substrate. For this technique, a
recombinant DNA is inserted within the coding sequence of an enzyme, β-galactosidase. Beta-galactosidase converts galactose into lactose. If a gene is inserted into this region, β-galactosidase will not be formed and therefore galactose will not be converted into lactose. This results in the inactivation of the enzyme. This is called as insertional inactivation. The presence of a chromogenic substrate causes non-transformed colonies to give blue colour. Presence of gene of interest results in the insertional inactivation of the galactosidase and the colonies therefore, do not produce any colour. These colonies can be inferred as recombinant colonies.
Insertional inactivation:
The inactivation of an enzyme due to the insertion of gene of interest in the region of DNA coding for the enzyme.
Vectors for cloning in plants:
Agrobacterium tumefaciens, a pathogen of several dicot plants is used as a vector for plants. It can deliver a piece of DNA known as ‘T-DNA’ to transform normal plant cells into a tumor and direct these tumor cells to produce the chemicals required by the pathogen. Gene of interest is inserted into T-DNA to transform plant cells with required gene. The tumor inducing (Ti) plasmid of Agrobacterium tumefaciens has now been modified into a cloning vector which is no more pathogenic to the plants. Cytokinin and auxin coding genes in plasmid acts as growth regulator. Opine catabolism gene codes for energy source. Right and left border are needed to transfer T-DNA into the required host plant cell.
 |
| Ti Plasmid |
Competent host
In order to allow bacterial cells to take up the DNA, bacterial cell should be made competent. This can be done by treating the cells with specific concentration of divalent ions such as calcium ions, which creates pores in the cell wall of the bacteria. Such bacteria are subjected to heat shock. In this method the calcium treated competent cells are kept in ice. They are then briefly incubated at 42◦C for 1-2 minutes and then immediately placed in ice. This forces the rDNA into the competent cell. Apart from this, DNA can be inserted into host cells using biolistics, microinjection, gene gun etc. Using microinjection, DNA can be directly inserted into the nucleus of the host cell. A high velocity microparticles of gold or tungsten coated with DNA is methodology used in biolistics.
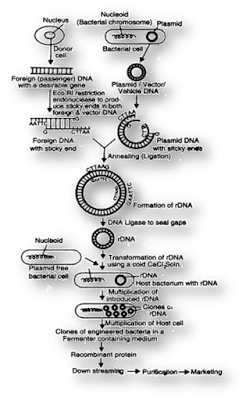
Processes of Recombinant DNA Technology
There are several steps involved in the process of recombinant DNA technology.
1. Isolation of the genetic material: To isolate the DNA, membranes needs to be broken down. Cells can be treated with lysozyme (in case of bacteria), cellulase (in case of plant cells), and chitinase (in case of fungus). Ribonucleases are used to remove the RNA whereas proteases are used to remove the proteins. After this, the pure DNA can be obtained through precipitation via ethanol. DNA is then obtained as fine threads in suspension.
2. Restriction digestion of the isolated DNA or Cutting of DNA: Agarose gel electrophoresis is used to check the progression of restriction digestion of the DNA. The gene of interest is now inserted into specific vector and joined via enzyme known as ligase. This forms a recombinant DNA molecule.
3. Amplification of gene of interest using PCR: Polymerase chain reaction (PCR) is used to amplify the target gene of interest. For this two sets of primers- forward primer and reverse primer is used. DNA polymerase enzyme is used to amplify the DNA. The most common polymerase used during PCR is Taq polymerase.
4. Insertion of recombinant DNA is into host cell or organism: Recipient cell is made competent to take up the recombinant DNA.
5. Expression of desired protein: The ultimate goal of recombinant DNA technology is to obtain desired protein of interest. The protein obtained is known as recombinant protein.
 |
| Recombinant DNA Technology |
Steps of recombinant DNA technology
To produce large quantities of recombinant protein, large vessels known as bioreactors are used. A bioreactor provides the optimal conditions for achieving the desired product by providing optimum growth conditions (temperature, pH, substrate, salts, vitamins, oxygen).
Basic parts of a bioreactor:
1. Agitator
2. Oxygen Control system
3. Foam control system
4. Temperature control
5. pH control
6. Sampling port
7. Inlet
8. Outlet
Bioreactors are mainly of two types: Stirred type and the sparger type
Stirring type bioreactor:
A stirrer is fixed to a bioreactor having a curved base to facilitate better mixing of the contents. It also improves aeration of the medium.
Sparger type bioreactor:
In this air is bubbled into the bioreactor from the base of the bioreactor. This bubbling of air results in mixing as well as aeration of the contents.
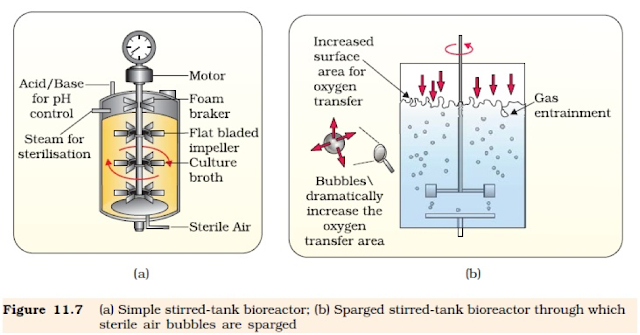 |
| Bioreactor |
Downstream Processing
The processes and methods involved in the separation and purification of the desired product are called as downstream processing. In case of drugs, the product needs to be suitably formulated and drug tested before being made available commercially.
 |
| Downstream Processing |
Thank you for reading. If you like our article then share with your friends.



